Microbial typing refers to characterization of an organism beyond its species level.
Applications of Microbial Typing:
Microbial
typing is an important tool for hospital microbiologists and epidemiologists. It
is used to determine the relatedness between different microbial strains of the
same species and thereby it helps to:
- Investigate outbreaks: All isolates tracked in an outbreak should belong to similar type.
- Determine the source and routes of infections.
- Trace cross-infection, i.e. transmission of health care-associated pathogens.
- Differentiate virulent strains from avirulent strains of same species.
- Differentiate between recurrence and infection with new strain.
- Evaluate the effectiveness of control measures.
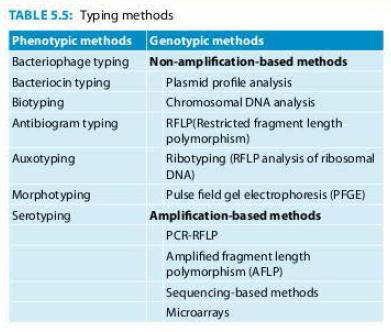
Classification of Microbial Typing: Typing methods are broadly classified as phenotypic and genotypic methods (Table 5.5).
Characteristic
of Typing Methods:
A
good typing method should have the following properties:
- Typeability: Ability of the method to type and generate a result for each isolate tested.
- Reproducibility: Ability to produce similar results when tested repeatedly in different laboratories.
- Discriminative power: Ability to generate distinct units of information making fine distinctions between the types at the subspecies level.
- Practicality: Ease of use and interpretation, cost and affordability.
In general,
genotypic methods are more reliable and have better reproducibility and
discriminative power than phenotypic methods, however they are expensive.
Phenotypic
Methods of
Bacteriophage Typing
Strains of an organism can be further differentiated into subspecies level based on their susceptibility to bacteriophages.
Phage typing is useful for the typing of:
Staphylococcus aureus
Salmonella typhi
Vibrio species
Brucella species
Corynebacterium diphtheria
Bacteriocin
Typing
Bacteriocin is an antibiotic like proteinaceous substance produced by one bacterium that inhibits other strains of the same or other closely related bacteria. Bacteriocin typing is based on the ability of a strain to produce particular bacteriocin which inhibits the growth of a set of selected indicator strains. It is done for:
Shigella sonnei ( colicin typing)
Klebsiella (klebocin typing)
Escherichia coli (colicin typing)
Proteus (proticin typing)
Pseudomonas (pyocin typing)
Biotyping
It refers to intra species classification based on different biochemical properties of the organism. It is used for:
- Corynebacterium diphtheriae: It is classified into gravis, intermedius and mitis.
- Vibrio cholerae O1 is classified into two biotypes-classical and El Tor
- Yersinia pestis.
Antibiogram
Typing
It classifies the organism into different groups based on their resistance pattern to different antimicrobials. Since antimicrobial susceptibility testing is routinely done in any hospital, this typing system provides the first clue to a microbiologist about outbreaks occurring in a hospital.
Auxotyping
This is a typing method based on nutritional requirement of the organism. This is done for Neisseria gonorrhoeae.
Morphotyping
This is based on different morphological appearances of the colonies in the culture media. This is done for Pseudomonas.
Serotyping
It refers to a typing method based on the antigenic property of an organism. This is the most widely used and the mot reliable phenotypic typing method. Serotyping is done for many organisms; important ones are given below.
- Streptococcus (Lancefield grouping; based on carbohydrate antigen).
- Based on capsular antigen-e.g. pneumococcus, meningococcus and Haemophilus influenzae.
- Based on somatic antigen- Escherichia coli, Shigella, Salmonella and Vibrio cholerae.
Genotypic
Methods of Microbial Typing
Plasmid
Profile Analysis
It is a method of determining a number and size of plasmids present in bacterial isolates. Plasmids produced by various strains in an outbreak are compared. First the plasmids are extracted from the bacterial cell and are then separated on agarose gel based on molecular weight followed by their detection by staining with ethidium bromide.
Restricted
Fragment Length Polymorphism (RFLP)
1. Digestion of DNA: This is done by using 2 or more restriction enzymes which cleave the DNA from a bacterial strain at different sites so that multiple DNA fragments are generated.
2. Southern blot to detect DNA fragments: The DNA fragments are separated by electrophoresis and transferred to a nitrocellulose membrane and then are detected by using specific DNA probes.
The
pattern of fragments generated by different strains tracked in an outbreak can
be compared to know the relatedness between the strains.
Ribotyping
Ribotyping
is a type of RFLP analysis which is done on chromosomal DNA coding for
ribosomal RNA.
Pulse
Field Gel Electrophoresis (PFGE)
PFGE is
considered as a gold standard method in epidemiological investigation of
pathogenic organisms. It has the following unique properties:
- Lysis: First, the bacterial suspension is loaded into an agarose suspension. This is done to protect the chromosomal DNA from mechanical damage by immobilizing it into agarose blocks. Then the bacterial cell is lysed to release the DNA. The agarose-DNA suspension is also known as plug mold.
- Digestion of DNA: The bacterial DNA is treated with rare cutting restriction enzymes so that it yields generation of less number of larger size DNA fragments (in contrast to frequent cutting restriction enzymes used in RFLP which produces large number of smaller fragments).
- Electrophoresis: The larger pieces of DNA are subjected to pulse field gel electrophoresis by applying electric current and altering its direction at a regular interval (in contrast to the conventional agarose gel electrophoresis done to separate the smaller fragments where the current is applied in a single direction).
- Analysis: The fragments generated by PFGE of various strains obtained during an outbreak are compared manually or by computer software BioNumerics.
The drawbacks
of PFGE are (!) It is labor intensive, (2) Requires many days to perform the
procedure, (3) Requires skilled personnel to interpret the results and (4) Requires
computer assisted analysis of banding patterns.
Amplified
Fragment Length Polymorphism (AFLP)
AFLP
uses the principle of performing RFLP of the bacterial DNA followed by PCR.
- The genomic DNA is digested by restriction enzymes, followed by use of adaptors to ligate to the sticky ends of the restriction fragments.
- PCR amplification of the restriction fragments is carried out by using primers complementary to both adaptor and restriction site sequences.
- The amplified fragments are separated and visualized on denaturing polyacrylamide gels.
Sequencing-based
Methods
The nucleotide sequence of a microbial gene can be obtained
by specially
designed equipment called sequencer. The
variability within the sequences of particular genes can be used to determine
the relatedness of bacteria. Sequence analysis can be done:
- At a single nucleotide (single nucleotide polymorphism or SNP analysis).
- Multiple genes (multilocus sequence typing or MLST).
- Whole genome sequencing.
Microarrays
Microarray
technology offers a wide range of analysis of simultaneous detection of multiple
gene products, such as antibiotic resistance determinants and virulence factors
whose identification can be useful for epidemiological investigations.
- The principle of the microarray is based on generating labelled cDNA or cRNA molecules that are subsequently hybridized to an arrayed series of thousands of microscopic spots with specific complementary oligonucleotides (probes).
- DNA microarrays have been used to measure changes in expression levels and to detect SNPs. It is also used for genotyping.

No comments:
Post a Comment