Gene transfer in bacteria can be broadly divided into-
- Vertical gene transfer (transmission of genes from parents to offspring during cell division)
- Horizontal gene transfer (transmission of genes from one bacterium to another neighbor bacterium)
Horizontal
gene transfer occurs in bacteria by several methods, such as:
- Transformation (uptake of naked DNA)
- Transduction (through bacteriophage)
- Lysogenic conversion
- Conjugation (plasmid mediated via conjugation tube)
TRANSFORMATION IN BACTERIA
Definition
Transformation
is a process of random uptake of free or naked DNA fragment from the
surrounding medium by a bacterial cell and incorporation of this DNA fragment
into its chromosome in a heritable form.
Natural
transformation has been studied so far only in certain bacreria-Streptococcus, Bacillus, Haemophilus, Neisseria, Acinetobacter and Pseudomonas.
Mechanism of Transformation in Bacteria
When
bacteria lyse, they release large amounts of dsDNA into the surrounding
environment. Their uptake depends upon the competency of the bacteria present
in the surroundings.
Competency
for Transformation
Competent
bacteria refers to the cells multiplying in log phase of cell division and
expressing certain transformation promoting factors called competence factors.
- Bacteria expressing competence factors (e.g. S. pneumoniae) can uptake any DNA fragment irrespective of source.
- But competence factors are not expressed by all bacteria that mediate transformation e.g. Haemophilus influenzae. In such case, the uptake of DNA occurs only from the closely related species.
The
transformation frequency of very competent cells is around 10-3 for
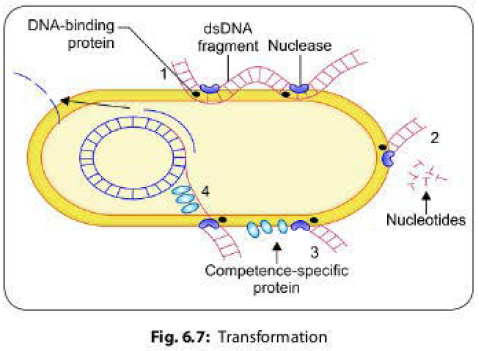
most genera. Steps involved in transformation are as follows (Fig 6.7):
1. A long dsDNA fragment comes in contact with a competent bacterium and binds to DNA-binding protein present on its surface and then it is nicked by a nuclease.
2. One strand is degraded by the recipient cell exonucleases.
3. The other strand associates with a competence specific protein and is internalized, which requires energy expenditure.
4. The single strand enters into the cell and is integrated into the host chromosome in place of the homologous region of the host DNA.
Griffith
Experiment
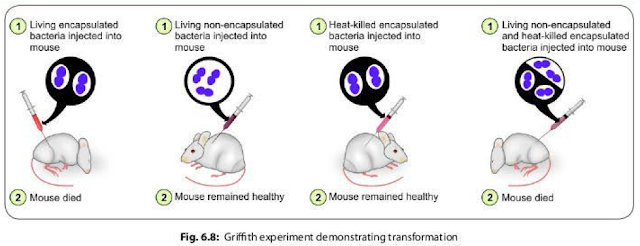
The
famous Griffith experiment (1928) on mice using pneumococci strains provided the
direct evidence of existence of transformation.
- Griffith found that mice died when they were injected with a mixture of live non-capsulated pneumococci and heat killed capsulated pneumococci strains. However, neither of which separately proved fatal to mice (Fig. 6.8).
- He stated that the live non-capsulated strains were transformed into the capsulated strains due to transfer of the capsular genes released from the lysis of the killed capsulated strains, which was confirmed later by Avery, Macleod and McCarty in 1944.
TRANSDUCTION IN BACTERIA
Definition
Transduction
is defined as transmission of a portion of DNA from one bacterium to another by
a bacteriophage (bacteriophage is a virus that infects and multiplies inside the
bacterium).
Mechanism
of Transduction in Bacteria
During
the transmission of bacteriophages from one bacterium to another, a part of the
host DNA may accidentally get incorporated into the bacteriophage and then gets
transferred to the recipient bacterium. This leads to acquisition of new characters
by the recipient bacterium coded by the donor DNA.
Bacteriophages
perform two types of life cycle inside the host bacteria.
1. Lytic or virulent cycle: Bacteriophage multiplies in host cytoplasm, produces large number of progeny phages, which subsequently, are released causing death and lysis of the host cell.
2. Lysogenic or temperate cycle: In contrast to virulent cycle, here the host bacterium is unharmed. The phage DNA remains integrated with the bacterial chromosome as the prophage, which multiplies synchronously with bacterial DNA. However, when the phage DNA tries to come out, it is disintegrated from host chromosome, comes out into the cytoplasm, and behaves as a lytic phage. It replicates to produce daughter phages, which are subsequently released by host cell lysis.
Types
of Transduction in Bacteria
Transduction
is of two types, either generalized or restricted.
Generalized Transduction
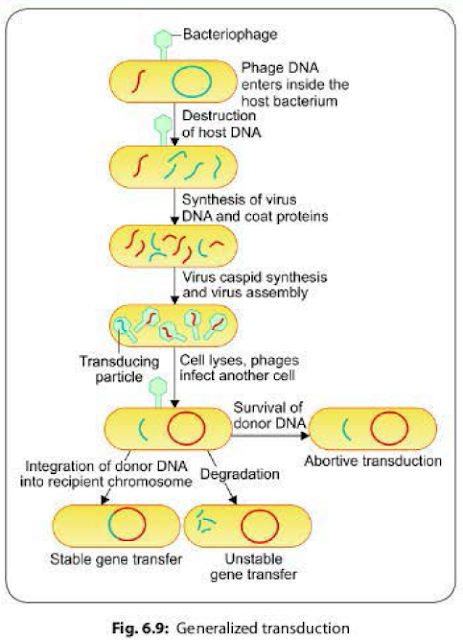
It involves
transfer of any part of the donor bacterial genome into the recipient bacteria.
Generalized transduction usually occurs as a result of defective assembly
during the lytic cycle of virulent and some temperate phages.
- Packaging errors may happen occasionally due to defective assembly of the daughter phages. Instead of their own DNA, a pan of host DNA may accidentally be incorporated into the daughter bacteriophages.
- The resulting bacteriophage (called transducing phage) often injects the donor DNA into another bacterial cell but does not initiate a lytic cycle as the original phage DNA is lost.
- The donor DNA may have three fates inside the recipient bacterium (Fig 6.9):
- Abortive transduction: About 70-90% of the transferred DNA is not integrated with the recipient bacterial chromosome, but often is able to survive and express itself. Such bacteria containing this non-integrated, transduced DNA are called abortive transductants.
- Stable gene transfer: The donor DNA gets integrated with recipient bacterial chromosome.
- Unstable gene transfer: In some cases, the donor DNA gets disintegrated by the host call enzymes.
Restricted or Specialized
Transduction in Bacteria
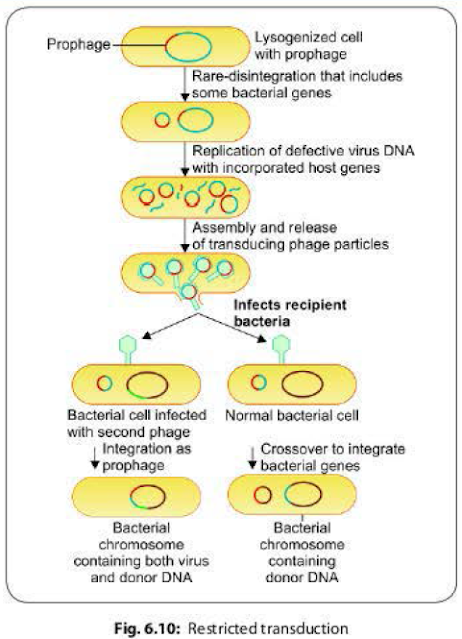
In contrast to generalized transduction, the restricted transduction is capable of transducing only a particular genetic segment of the bacterial chromosome that is present adjacent to the phage DNA.It occurs as a result of defect in the disintegration of the lysogenic phage DNA from the bacterial chromosome.
- Restricted transduction has been stuthed intensively in the 'lambda' phage of E. coli.
- When a prophage (i.e. lysogenic bacteriophage is integrated with the bacterial chromosome) leaves the host chromosome, portions of the bacterial chromosome present adjacent to the phage DNA may get wrongly excised along with it.
- Such transducing phages carrying a part of bacterial DNA in addition to their own DNA, when infect another bacterium, the transfer of the donor DNA takes place in two ways (Fig. 6.10).
- Crossover between the donor DNA and a part of recipient DNA- leads to integration of the donor DNA into the recipient chromosome and a part of recipient DNA into the phage DNA.
- The entire transducing genome (ie. phage DNA + donor DNA) acts as a prophage and gets integrated to the recipient chromosome. This occurs if the recipient bacterium is already infected by another helper bacteriophage.
Role of Transduction in Bacteria
In addition
to chromosomal DNA, transduction is also a method of transfer of episomes and
plasmids.
- Drug resistance: Transduction may be a mechanism for the transfer of bacterial genes coding for drug resistance; for example, plasmid coded penicillin resistance in staphylococci.
- Treatment: Transduction has also been proposed as a
method of genetic engineering in the treatment of some inborn metabolic
defects.
LYSOGENIC
CONVERSION IN BACTERIA
During
the temperate or lysogenic life cycle, the phage DNA remains integrated with
the bacterial chromosome as prophage, which multiplies synchronously with the bacterial
DNA.
- The prophage acts as an additional chromosomal element which encodes for new characters and is transferred to the daughter cells. This process is known as lysogeny or lysogenic conversion.
- Imparts toxigenicity to the bacteria: Phage DNA may be responsible for bacterial virulence by coding for their toxins production. For example, in Corynebacterium diphtherie, the diphtheria toxin is coded by a lysogenic phage DNA which is integrated with the bacterial chromosome. Elimination of the phage from a toxigenic strain renders the bacterium non-toxigenic.
Phage Coded Toxins
Bacterial toxins that are coded by lysogenic phages include:
- Diphtheria toxin
- Cholera toxin
- Verocytotoxin
of E.coli
- Streptococcus pyrogenic exotoxin (SPE)-A and C
- Botulinum toxin C and D
- In lysogenic conversion, the phage DNA itself behaves as the new genetic element in contrast to transduction where the phage acts only as a vehicle carrying bacterial genes.
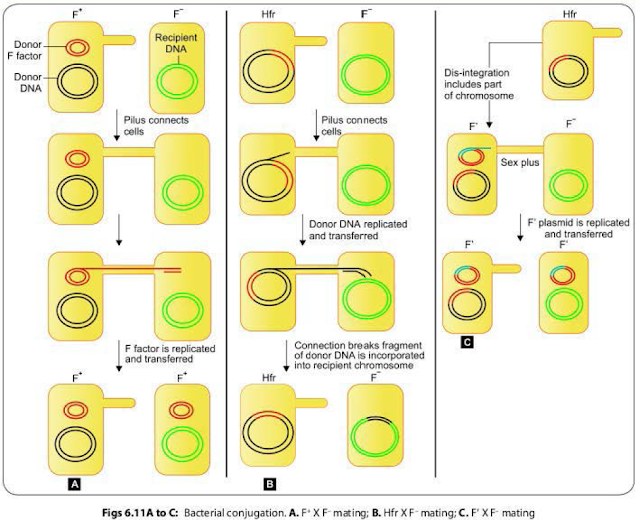
CONJUGATION IN BACTERIA
Conjugation
refers to the transfer of genetic material from one bacterium (donor or male)
to another bacterium (recipient or female) by mating or contact with each other
and forming the conjugation tube. It was discovered first by Lederberg and
Tatum (1946).
F+ X F-
Mating
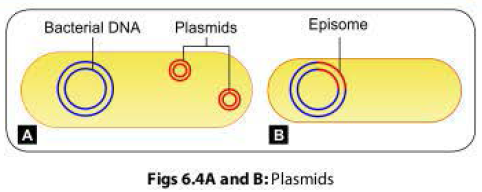
The F+ cell (also called as the donor or the male bacterium) contains a plasmid called as F factor or fertility factor.The bacteria lacking the F factor are called as female or recipient bacteria or F- cell.
- F factor is a conjugative plasmid; carries genes that encode for the formation of sex pilus (that helps in conjugation) and self plasmid transfer.
- The F pilus brings the donor and nearby recipient cells close to each other and form a conjugation tube that bridges between the donor and recipient cells (Fig. 6.II A).
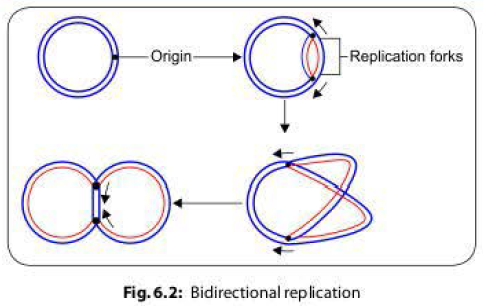
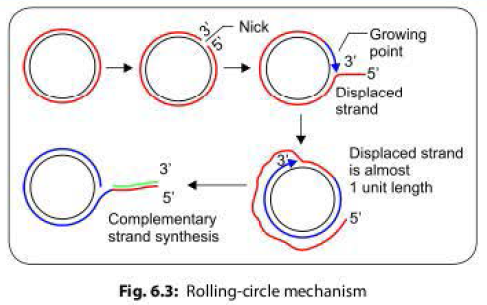
- During conjugation, the plasmid DNA replicates by the rolling-circle mechanism, and a copy moves to the recipient bacterium through the conjugation tube. Then, in the recipient, the entering strand is copied to produce complete F factor with ds DNA.
- As a result, recipient (P) becomes (F') cell and can in turn conjugate with other (F-) cells. Therefore, it is said that this character of maleness (F') in bacteria is transmissible or infectious.
- During F' X F-conjugation, chromosomal genes from donor bacterium may rarely be transferred along with F factor. Here, though the donor chromosomal gene may undergo recombination with the recipient chromosome; but with a lower frequency.
HFR Conjugation
F
factor being a plasmid, it may integrate with bacterial chromosome and behave
as episome.
- Such donor cells are able to transfer chromosomal DNA to recipient cells with high frequency in comparison to F' cells, therefore, named as HFr cells (high frequency of recombination).
- During conjugation of HFr cell with an F- cell, only few chromosomal genes along with a part of the F factor get transferred. Connection between the cells usually breaks before the whole genome is transferred.
- As the entire F factor does not get transferred, hence following conjugation, F- recipient cells do not become F+ cells (Fig. 6.118).
F’ Conjugation
The
conversion of an F+ cell into a Hfr cell is reversible.
- When the F factor reverts from the integrated to free state, it may sometimes carry with it some chromosomal DNA from adjacent site of its attachment. Such an F factor carrying some chromosomal DNA is named as F' factor (F prime factor).
- When F' cell conjugates with a recipient (F-, it transfers the host DNA incorporated with it along with the F factor. The recipient becomes F' cell. This process is called sexduction. (Fig. 6.11 C).
Conjugation
plays an important role in the transfer of plasmids coding for antibacterial drug
resistance(resistance transfer factor (RTF) and bacteriocin production
(Colicinogenic (Col) factor].
Colicinogenic (Col) Factor
The
bacteriocin production in bacteria is plasmid coded which may be transferred by
conjugation. Such plasmids are called as the col factors. Bacteriocins are the antibiotic-like
substances produced by one bacterium that inhibit other bacteria. Bacteriocins
produced by coliform bacteria are called as colicin. Bacteria other than coliforms
also produce similar kind of substances e.g. pyocin by Pseudomonas, diphthericin by Corynebacterlum diphtheriae.
Resistance Transfer Factor (RTF)
- Conjugation is also an important method of transfer of plasmids coding for multiple drug resistance among bacteria.
- R factor (or the resistance factor) is a plasmid which has two components. (R factor = RTF+ r determinants).
- Resistance transfer factor (RTF): It the plasmid responsible for conjugational transfer (similar to F factor)
- Resistance determinant (r): R factor can have several r determinants and each r determinant coding for resistance to one drug.
- Sometimes, the R factor dissociates and both RTF and the r determinants exist as separate plasmids. In such cases, the resistance is not transferable though the host cell remains drug resistant.
- In addition to r determinants, the RTF can also attach to other genes; for example, genes coding for enterotoxin and hemolysin production in some enteropathogenic E.coli.
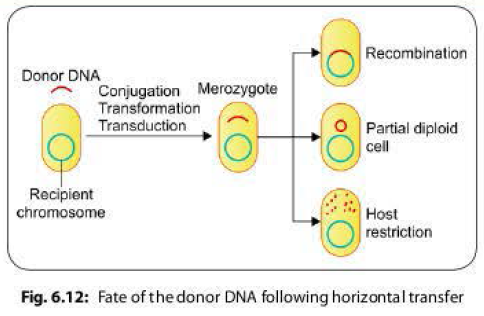
Fate of the Donor DNA Following Horizontal Transfer
Following
horizontal gene transfer by any of the methods described above, the donor DNA enters inside the
recipient cell, and remains in the cytoplasm temporarily. At this stage, the
recipient cell is called merozygote. The donor DNA has one of the following
fate inside the recipient cell (Fig. 6.12).
- Recombination: The donor DNA integrates with the recipient chromosome either as a replacement piece (usually occurs in transformation) or as an extra piece.
- Partially diploid cells: The donor DNA persists outside the host chromosome and the host cell becomes partially diploid for a portion of the genome that is homologous to the donor DNA. Such cells may or may not replicate to produce a clone of partially diploid cells.
- Host restriction: The host cell nucleases may degrade the donor
DNA if it is not homologous to any part of bacterial chromosome.
BACTERIAL RECOMBINATION
Recombination
takes place between the donor DNA and recipient chromosome in two ways: General
recombination and site specific recombination.
General or Homologous Recombination in Bacteria
It is
the most common form, can occur at any place on the recipient's chromosome in
general and occurs between DNA of similar sequences (homologous).
- Rec genes: The rec genes present in recipient's chromosome and their produces, such as the recA proteins are crucial to bring-out recombination.
- Crossing-over: The donor and recipient DNA strands breakage takes place followed by their reunion by crossing-over of strands.
- The Holliday model has been put forward (named after Robin Holliday, 1964), to describe the process of reunion.
Reciprocal vs Non-reciprocal
General Recombination
Most
of the donor DNA fragments entering into the recipient cell by horizontal gene
transfer are double stranded; except those transferred by transformation which
are single stranded. General recombination can be of two types:
- Reciprocal exchange: In most cases (except in transformation), the general recombination usually involves a reciprocal exchange between a pair of homologous DNA sequences between donor and recipient strands.
- Nonreciprocal exchange: In bacterial transformation, a nonreciprocal form of general recombination takes place. The single strand of donor DNA is inserted into the host chromosome (by replacing a piece of host chromosome) to form a stretch of heteroduplex DNA.
Site Specific Recombination
In
restricted transduction, the integration of bacteriophage DNA into bacterial
chromosome is site-specific.
- The donor DNA is not homologous with the chromosome it joins, and
- The enzymes responsible for this event are specific for the particular bacteriophage and its host bacterium.
TRANSPOSITION IN BACTERIA
Transposons
or transposable elements are the bacterial genes that are capable of intracellular
transfer between chromosome to chromosome, plasmid to plasmid, and chromosome
to plasmid or vice versa and the process of such intracellular transfer of transposons
is called as transposition. As transposons move around the genome in a
cut-and-paste manner, they are also called jumping genes or mobile genetic elements.
- Transposition does not require any DNA homology between transposon and the site of insertion. It is, therefore, different from recombination.
- Unlike plasmids, transposons are not self replicating and are dependent on chromosomal or plasmid DNA for replication.
- Transposons were first discovered in the 1940s by Barbara McClintock during her stuthes on maize genetics for which she won the Nobel prize in 1981.
- Transposons are also discovered in the virus and in eukaryotic genome.
Types of Transposons in Bacteria
Insertion Sequence
Transpason
The
simplest form of transposon is an insertion sequence.
It is
about 1-2 kilo base pairs (kbp) in length and consists of a transposase gene
(that helps in transposition) which is flanked at both the ends by inverted
repeat sequences of nucleotides, i.e. nucleotide sequences complementary to
each other but in the reverse order (Figs 6.13A and B).
Because
of this feature, each strand of the transposon can form a single-stranded loop
carrying the transposase gene, and a double stranded stem formed by hydrogen bonding
between the terminal inverted repeat sequences.
Composite Transposon
They
are larger transposons carrying additional genes, such as genes coding for
antibiotic resistance or toxin production in the center and both the ends are
flanked by insertion sequences that are identical or very similar in sequence
(Fig. 6.13C).
GENETIC ENGINEERING IN BACTERIA
Genetic engineering refers to deliberate
modification of an organism's genetic information by directly altering its nucleic
acid genome. Genetic engineering is accomplished by a precise mechanism known
as recombinant DNA technology.
The gene coding for any desired
protein is isolated from an organism, and then inserted into suitable vector,
which is then cloned in such a way that it can be expressed in the formation of
specific (desired) protein.
Recombinant DNA Technology
The procedure of recombinant DNA technology
involves the following steps:
1. Treatment with restriction enzyme: The DNA from the microorganism is extracted and then is cleaved by enzymes called restriction endonucleases to produce mixture of DNA fragments.
2. Southern blot: The fragment containing the desired gene is isolated from the mixture of DNA fragments.
This is done by:
- Electrophoresis: DNA fragments are electrophoretically separated by subjecting to agar gel electrophoresis.
- Transfer to nitrocellulose membrane: The separated DNA fragments are transferred from the gel to a nitrocellulose membrane.
- Detection of desired gene: The DNA fragment containing the desired gene is detected adding a specific DNA probe, complementary to the gene of interest.
- Isolation: The band containing the desired gene is isolated by DNA extraction and then, is subjected to electrophoresis in a different gel.
3. Recombination with a vector: The isolated DNA fragment is annealed with a vector by DNA ligase enzyme.
4. Introduction of the vector into bacteria: Thee vector is introduced into bacteria usually by transformation (injecting by electroporation) and rarely by phage vector by transduction.
5. Cloning: Culture of the bacteria containing the desired gene followed by expression of the gene products yields a large quantity of desired protein.
Applications of Genetic Engineering
- Production of vaccines: Preparation of certain vaccines is done by DNA recombination technology by producing the desired antigen that can be used as immunogen in vaccine, against which the protective antibody will be produced, e.g. vaccines for hepatitis B and human papilloma virus.
- Production of antigens used in diagnostic kits: llie antigens used in diagnostic techniques for antibody detection (e.g. ELISA) are prepared by DNA recombinant technology.
- Production of proteins used in therapy: Genetic engineering has also been used for the production of proteins of therapeutic interest. These include human growth hormone, insulin, interferons, interleukin-2, tumor necrosis factor, and factor VIII.
- Transgenic animals: Recombinant DNA technology can be used to artificially introduce a foreign DNA into the genome of animals. The process is called transfection and the recombinant animals produced in this way are named transgenic or genetically modified organisms. Transgenic mice are available for a variety of biotechnological applications.
- Gene therapy: Genetic diseases can be cured by replacing the defective gene by introducing the normal gene into the patient.
Vector
A vector is a small piece of DNA, into
which a foreign DNA fragment can be inserted and that can be stably maintained in
an organism and used for cloning purposes. There are four major types of vectors,
such as:
- Plasmids
- Bacteriophages
- Cosmids
- Artificial chromosomes, such as bacterial/yeast artificial chromosomes
NUCLEIC ACID PROBE
Nucleic acid probes are radiolabeled or
fluorescent labeled pieces of single stranded DNA or RNA, which can be used for
the detection of homologous nucleic acid by hybridization.
- Hybridization is the technique in which two single strands of nucleic acid come together to form a stable double stranded molecule.
- There are two types of nucleic acid probes- DNA probes (hybridizes with DNA) and RNA probes (hybridizes with RNA)
- Nucleic acid probes are used to detect the specific nucleic acid either-
- Directly in the clinical sample or
- Following amplification of small quantity nucleic acid present in the clinical sample (e.g. in real lime PCR) or
- Following enzymatic digestion of the extracted nucleic acids so that it detects only the specific DNA fragment from the mixture (e.g. in Southern blot).
BLOTTING TECHNIQUES
A blot, in molecular biology refers to
a method of transferring DNA, RNA, or proteins, from gel onto a carrier (e.g.
nitrocellulose membrane), followed by their detection by using specific nucleic
acid probes (for DNA or RNA detection) or enzyme immunoassay (for protein detection).
There are various blotting techniques:
- Southern blot is used to detect DNA
- Northern blot is used to detect RNA
- Western blot is used to detect proteins
- Eastern blot: It is a modification of Western blot, used to analyze proteins for post-translational modifications using probes that may detect lipids, carbohydrate, phosphorylation or any other protein modification.